Help
Introduction
Integrated Traditional Chinese Medicine, ITCM, is a user-friendly information platform that extensively curates mass scientific research data related to traditional Chinese medicine (TCM) and include three main modules: TCM literature (CMTP) MODULE, TCM small molecule expression profile (SMEP) module and TOOL module
➢ SMEP module
The SMEP module collects and analysze the largest uniform high-throughput sequencing datasets, including 1488 high-quality pharmacological transcription profiles of 496 active TCM ingredients. All profiles were obtained by our team through standard whole-transcriptome sequencing procedures of MCF-7 cell lines. Notably, profiles of 424 of TCM ingredients were firstly obtained. A standard and comprehensive analysis pipeline was conducted, including traditional analysis such as differential expression analysis, functional enrichment analysis, connectivity map (CMap) analysis. and newest analysis, gene set enrichment analysis (GSEA), gene set variation analysis (GSVA), transcription factors enrichment analysis (TFEA).In addition, gene signatures obtained from ingredients are upload to CMap and we clustered CMap results to obtain a heatmap exhibiting the largest pharmacological spectrum of TCM ingredients.
➢ TCM-Query
This module innovatively deploys advanced mining strategy in database to explore the relationship between specific geneset (SG, which can represent ingredient or disease) and profiles. See Q&A (What are the algorithms used in TOOL?) below for strategy description
➢ CMTP MODULE
The CMTP MODULE integrated and manually revised TCM data from:
● Mainstream TCM databases (SYMMAP, TCMSP, ETCM, NPASS, CMAUP, TCMIO, HERB, TCM-ID)
● Pharmacopeias of various countries, including the latest edition of Chinese Pharmacopoeia (2020), European Pharmacopoeia (10.0), and Herbal Medicines Compendium of The United States Pharmacopeia (December 2020).
● Kampo (Japanese TCM formulae from Journal Current Kampo )
● Additional TCM information from the literature database(Pubmed, CNKI)
● Official recommendation of formulae for COVID-19
Given that animal medicine and mineral medicine also play a vital role in TCM, we manually collected many pieces of information about animal medicine, mineral medicine, fungus medicine, which makes up for the deficiency of the current mainstream database. In this module, it includes 25857 formulae, 8454 herb+, 43430 ingredients, 18851 targets and 11180 disease.

Meanwhile, based on the ADMET properties of drugs predicted by an advanced AI algorithm, we inferred therapeutic effects for all the TCM ingredients.
To our knowledge, this module contained the most comprehensive data resource about TCM.
User Manual
Search data
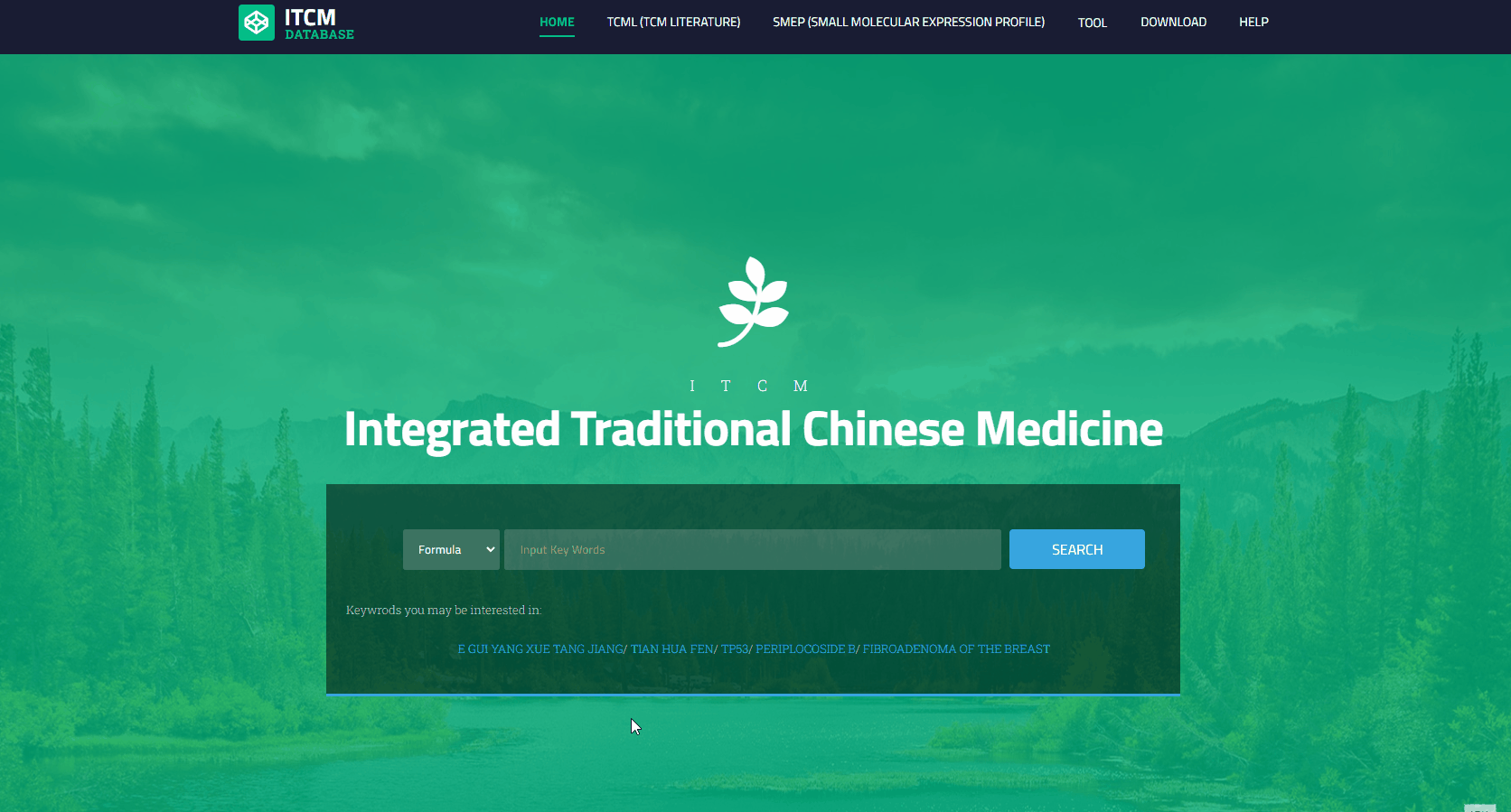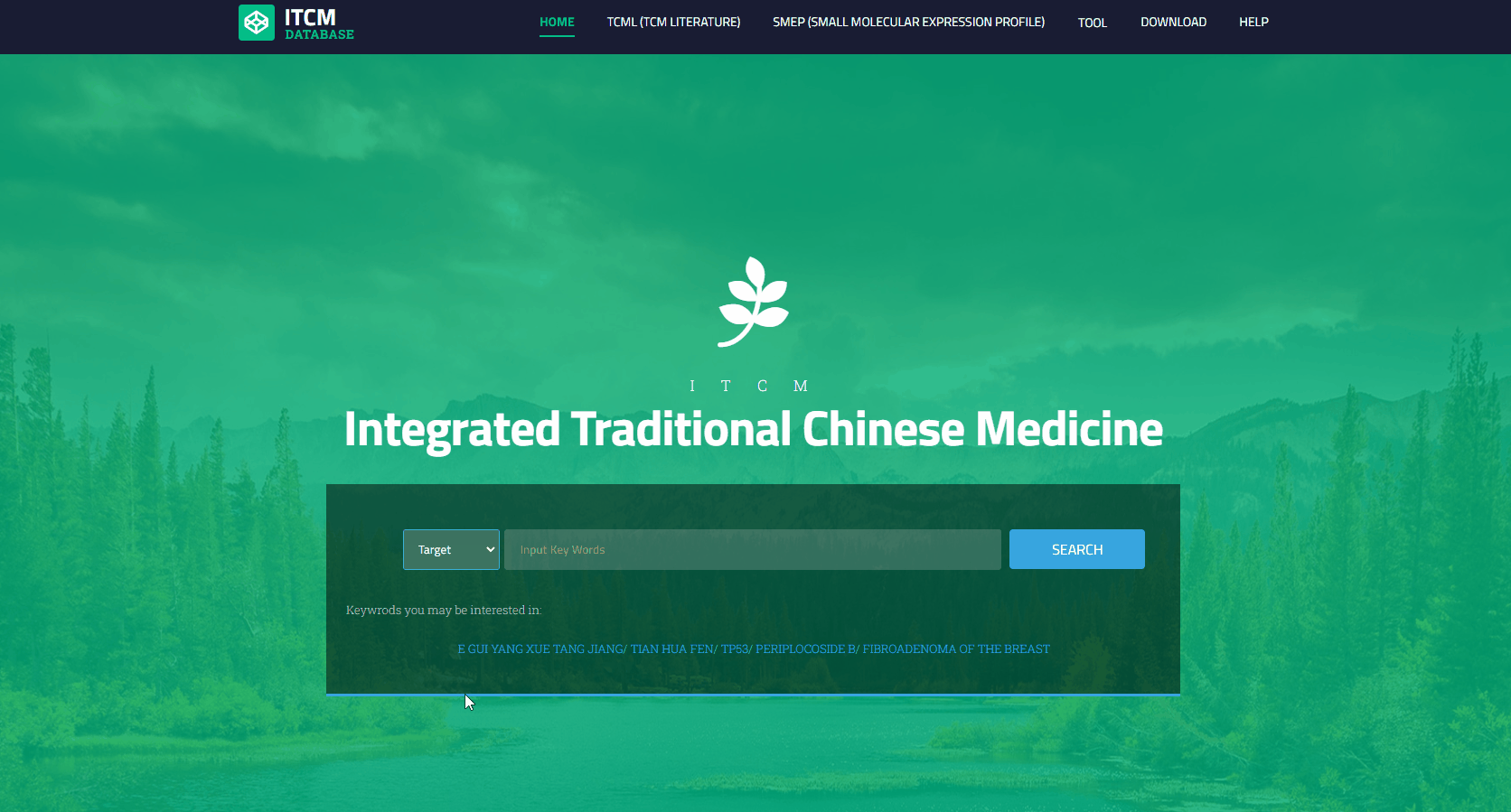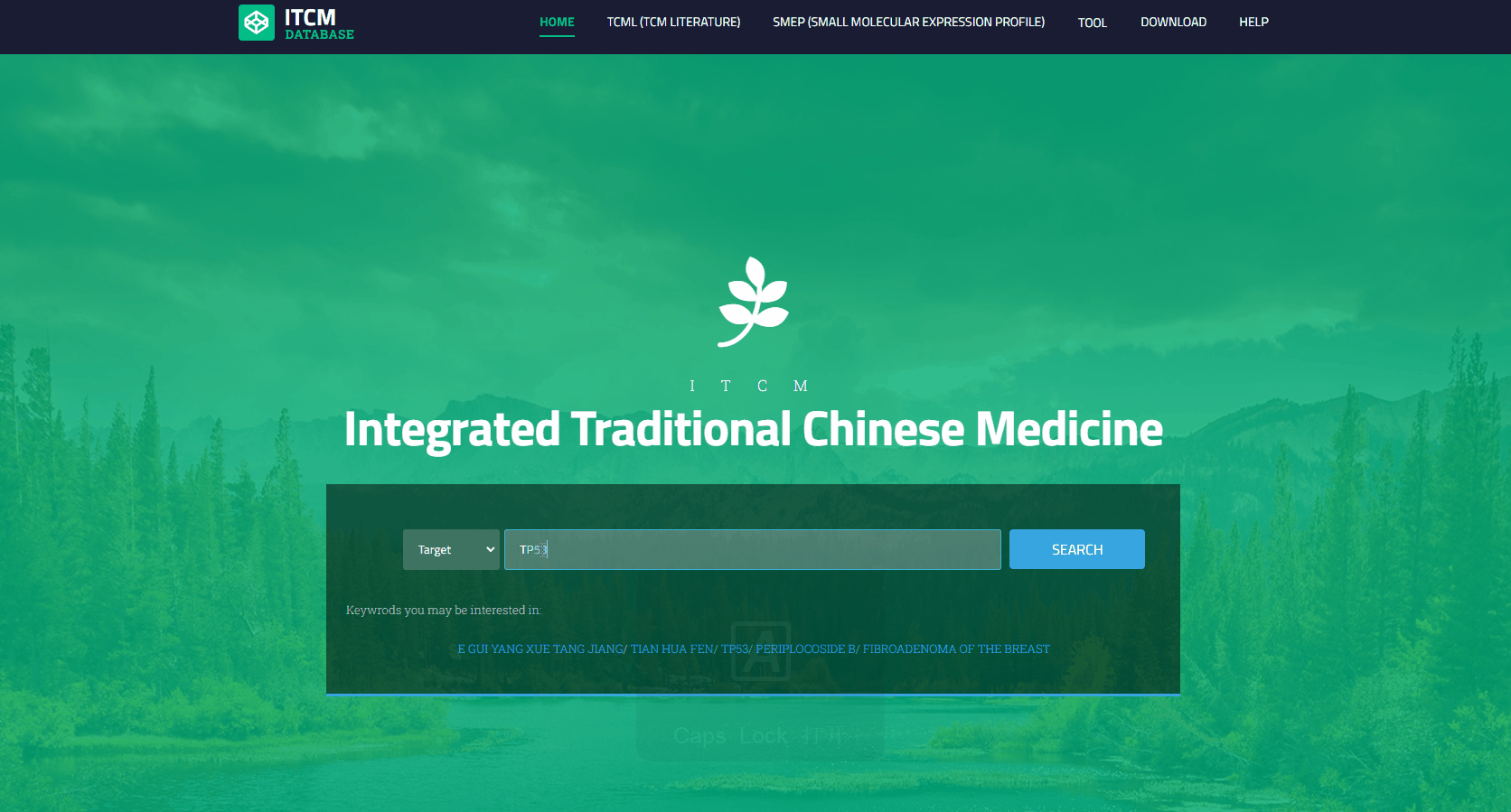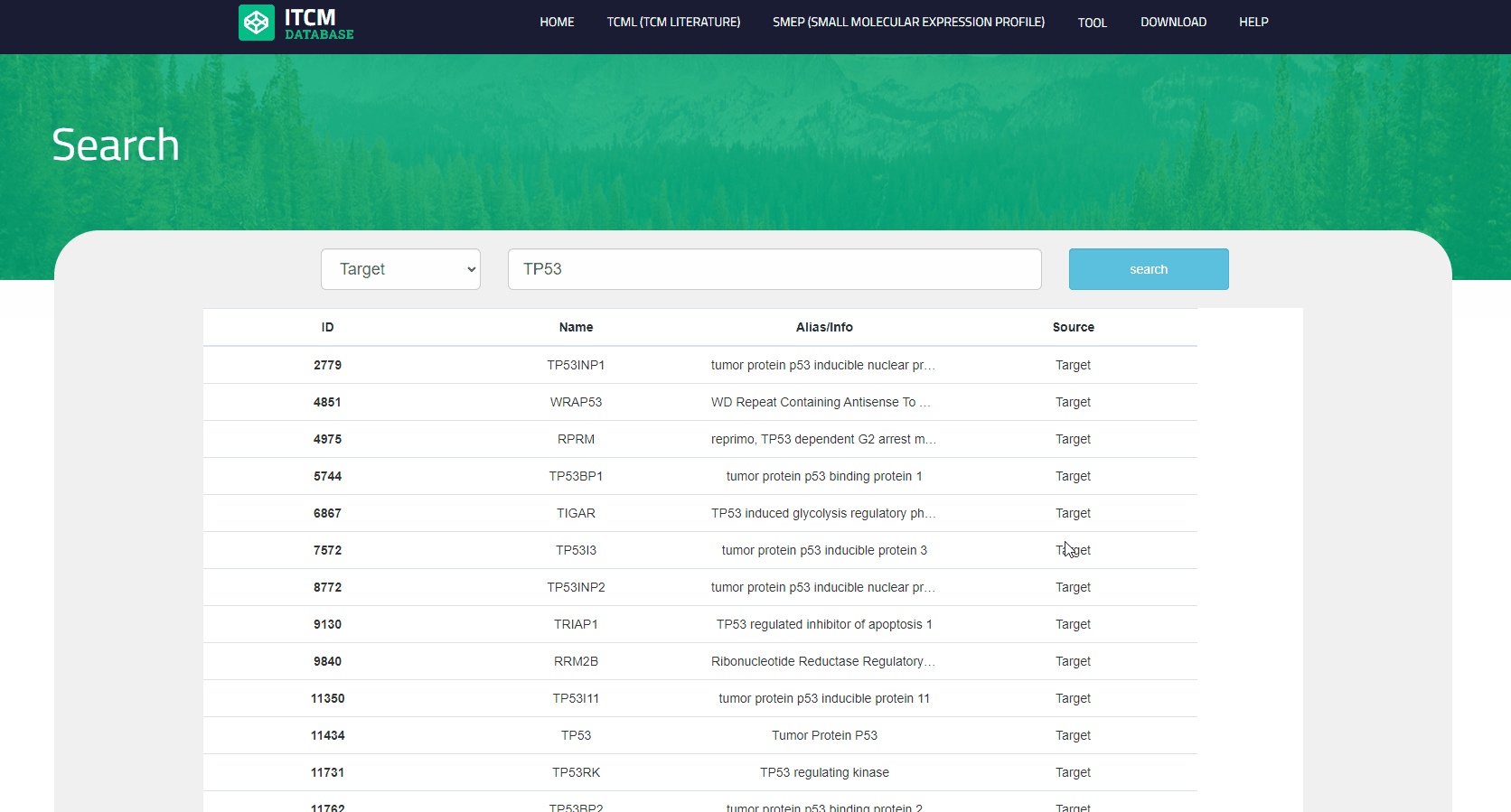
Users can search the database by inputting interesting queries in the search box at the center of the main page, then the web page returns a checklist. The default output results include all sources(formula, herb, ingredient, target, disease, profile), and the user can also focus on a specified source by clicking the dropdown box before search.
Notably, search in all fields may take a bit long time.
Browse data
User can browse data by clicking the button"CMTP" or "SMEP" on the banner of main page.

For literature data in CMTP MODULE
Users can browse the database by clicking the "CMTP" button on the navigation bar and select one category on the ribbon menu to view the records in a checklist. For each item, column names from left to right indicate ID, name, Alias/Info, source, and user can click any of them to the detailed page.
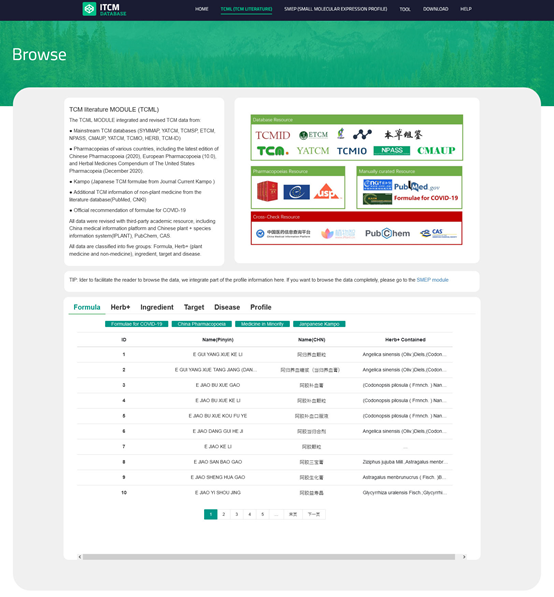
For Profiles in SMEP MODULE
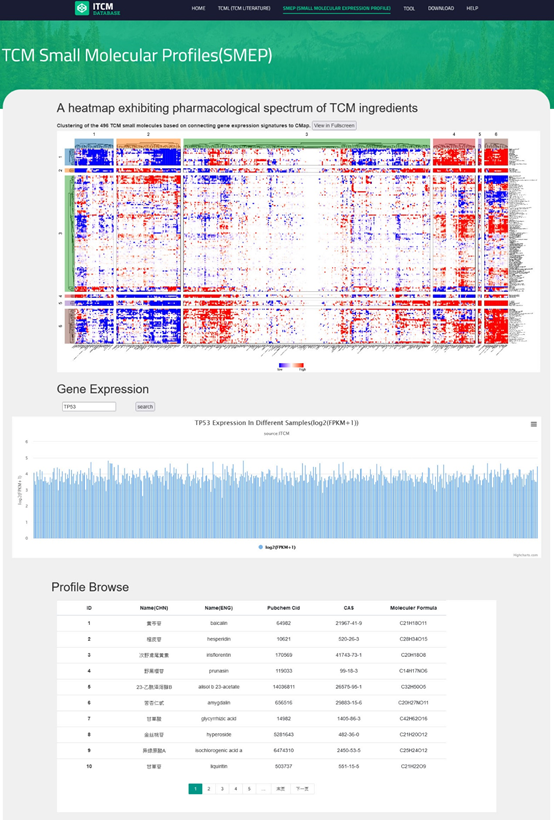
Users can browse the SMEP module by clicking the "SMEP" button on the navigation bar, and the main page of the profile contains 3 parts:
● a clustered heatmap for all connectivity map results over TCM small molecules
● user can input interesting genes in the search box to get an expression histogram across all profiles(mean expression of ingredient sample).
● in addition, at the bottom is an info table just like on the "CMTP" page and the user can click any of them to the detailed page.
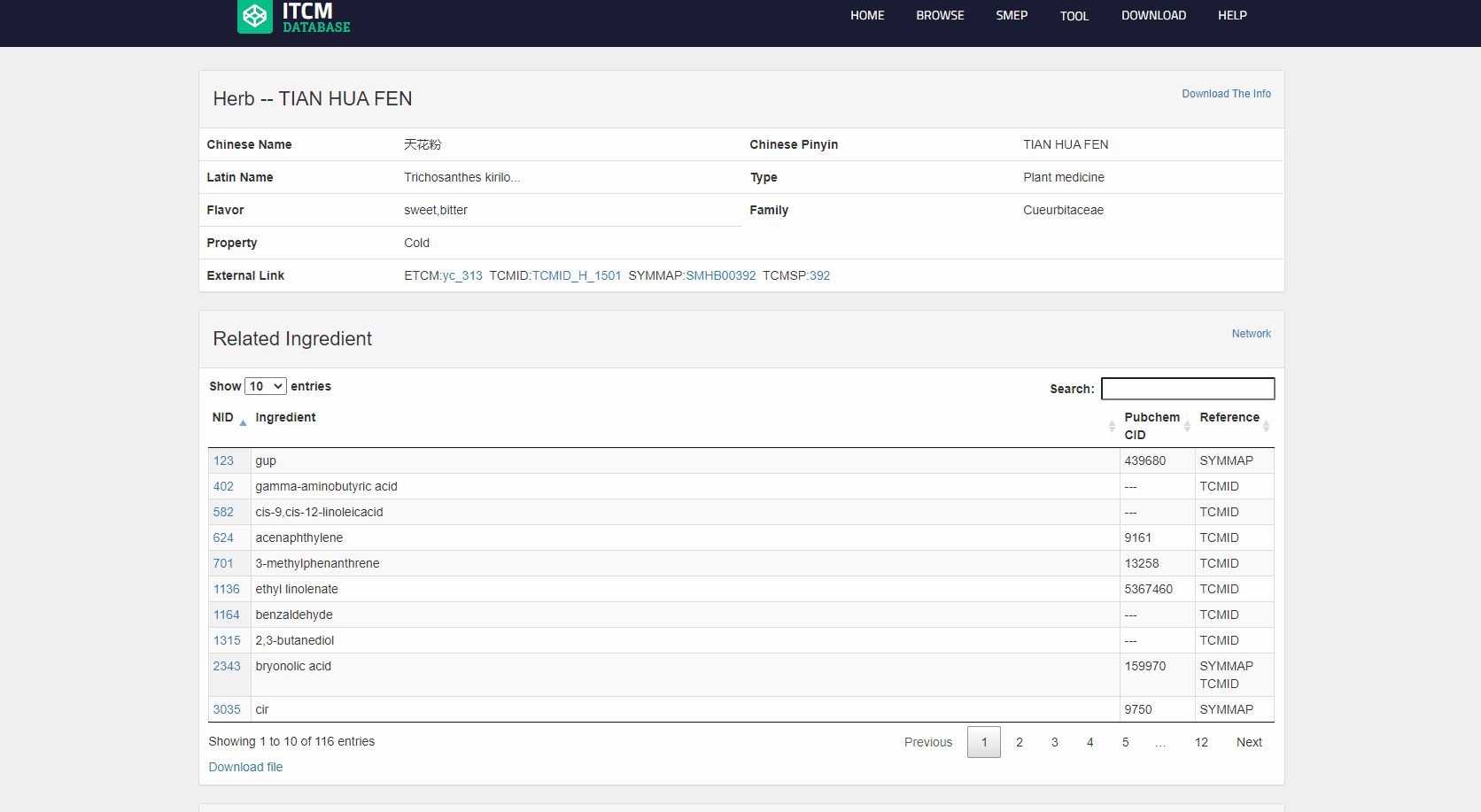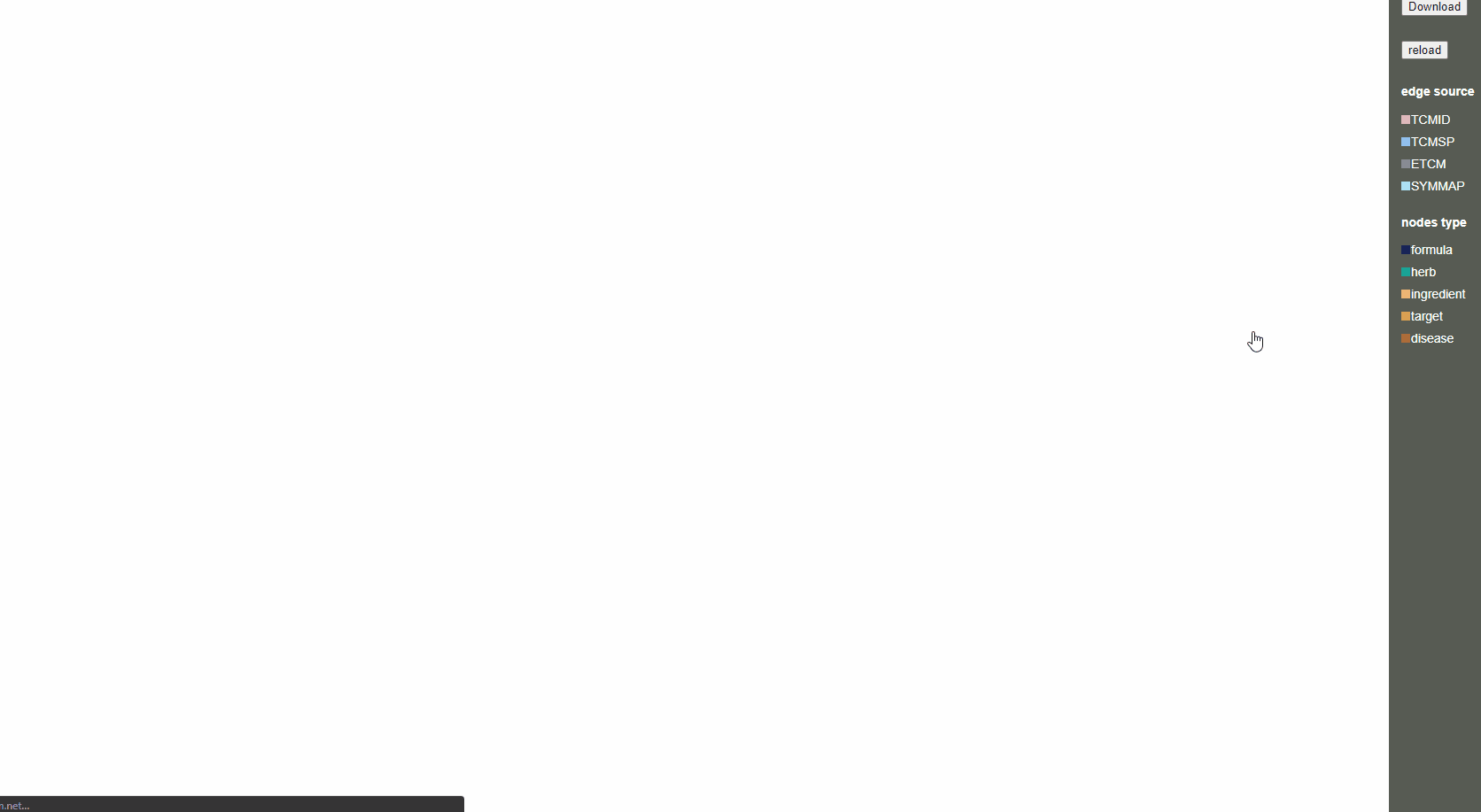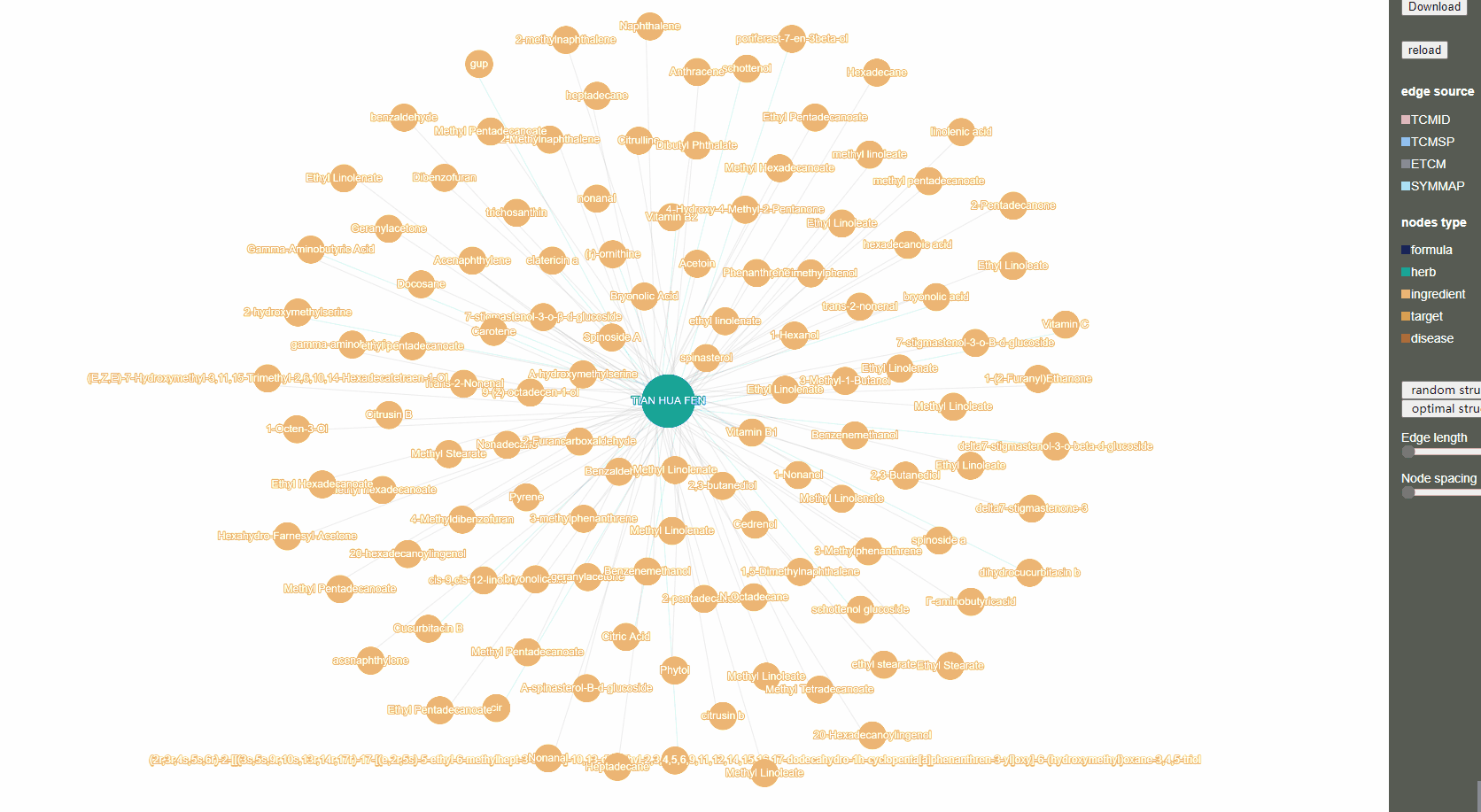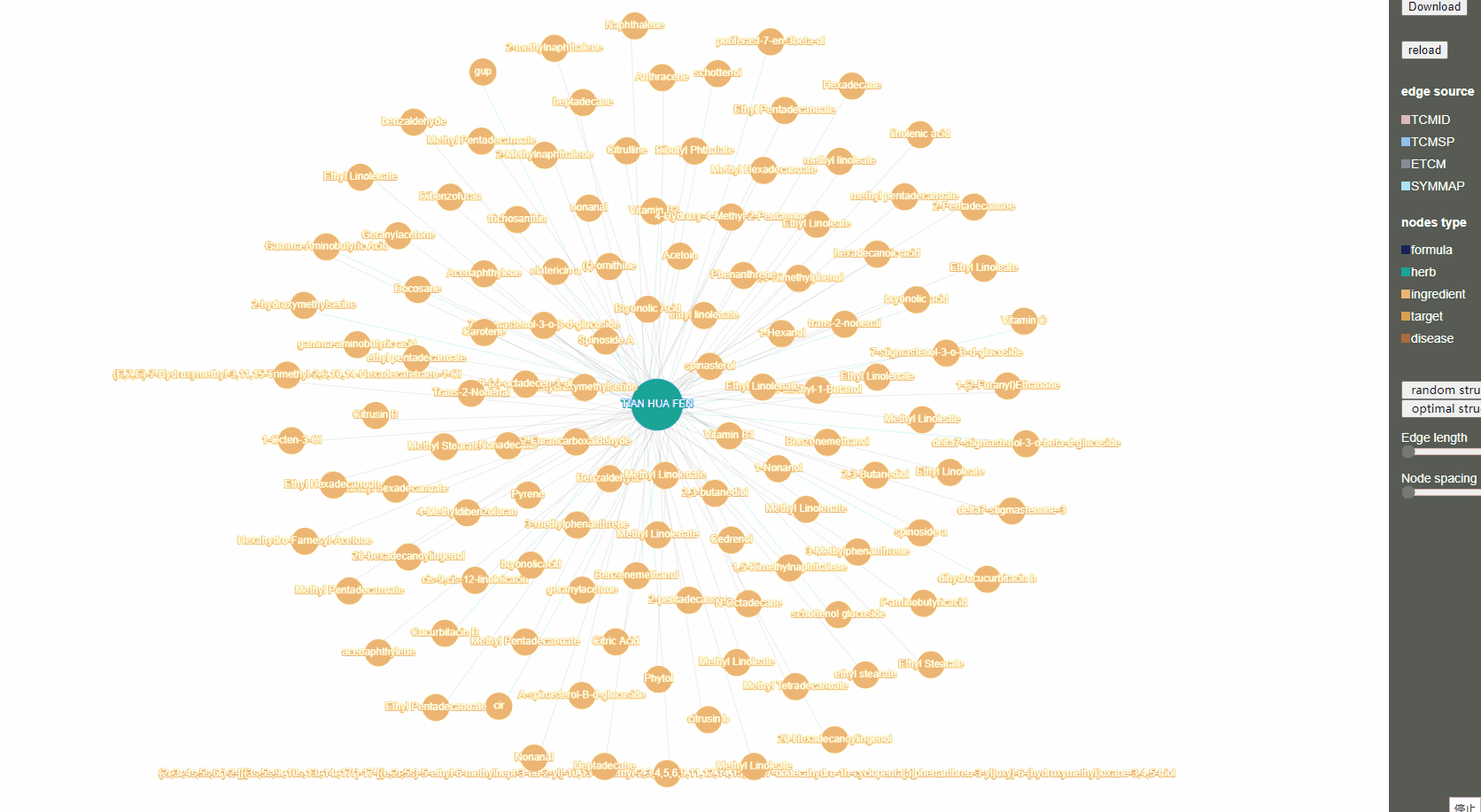
View the detailed page for each item in CMTP module
Users can enter the details page from the ‘CMTP’ and search results page by clicking the hyperlink of a certain item. the information cotained in item is summarized in the table below:
| source | Information contained | tips | Example link |
|---|---|---|---|
| Formula | Name(CHN, ENG) Brief in medical term External link Related Herbs Statistical Related Disease* Enrichment analysis |
* EASE score <0.01 | http://itcm.biotcm.net/detail/formula.html?nId=2 |
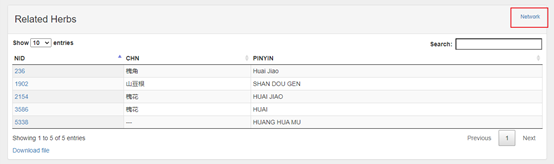
| Herb | Name(CHN, ENG) Brief in medical term External link Related Formula Related Ingredients Statistical Related Disease* Enrichment analysis |
* EASE score <0.01 | http://itcm.biotcm.net/detail/herb.html?nId=21 |
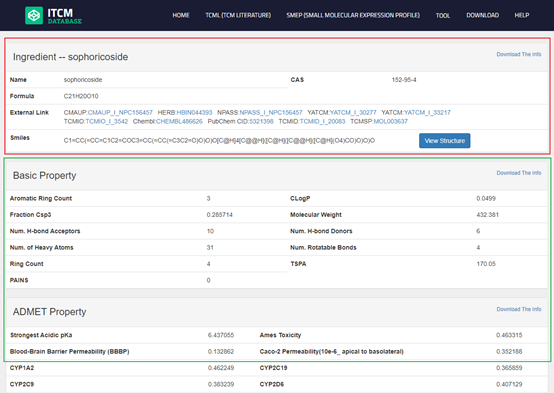
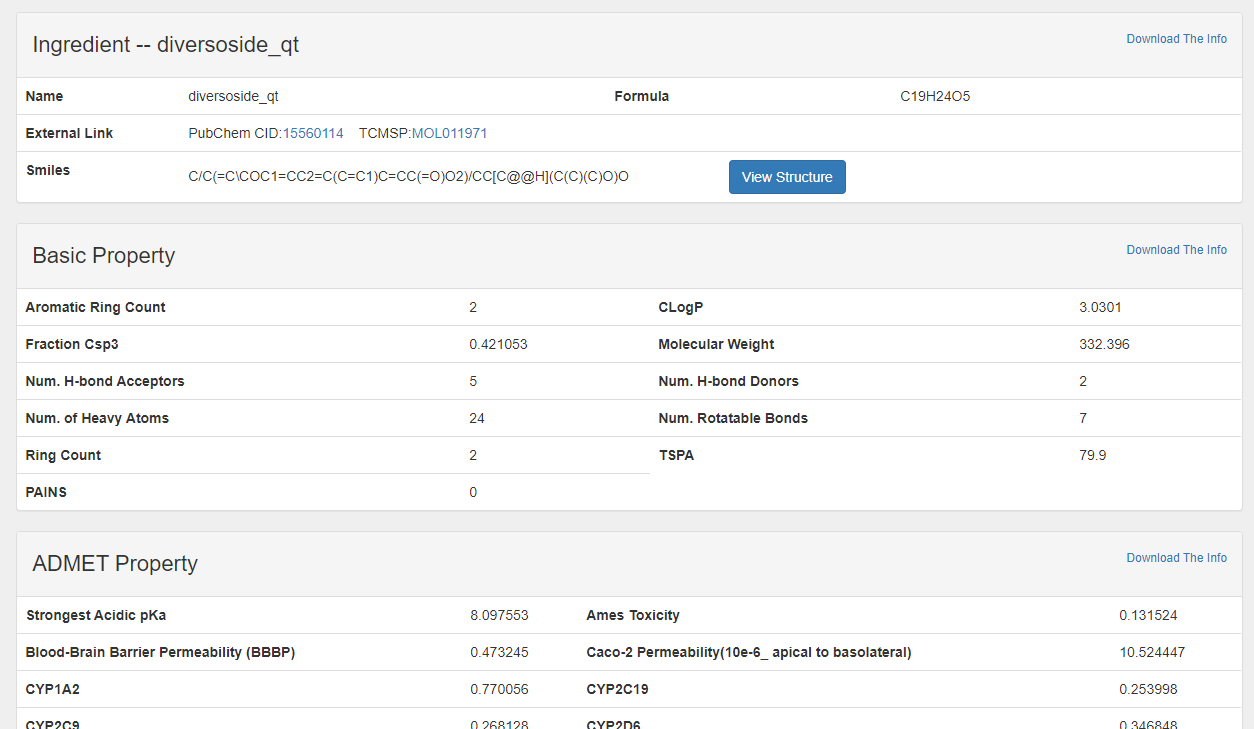
| Ingredient | Name(CHN, ENG) Brief in Chemical term Basic Property** ADMET Property** Drug-like inference*** External link Related Herbs Related Target Statistical Related Disease* |
* EASE score <0.01 ** predicted by iDrug *** see Q&A |
http://itcm.biotcm.net/detail/ingredient.html?nId=9148 |
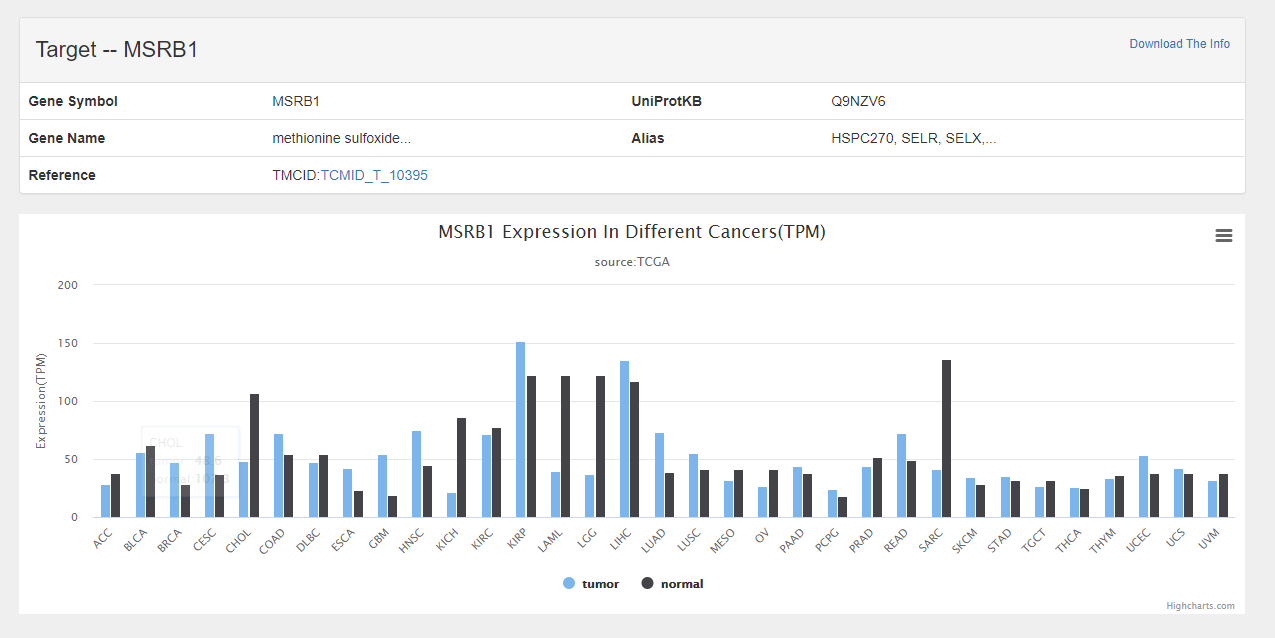
| Target | Name Gene Symbol External reference External link Related ingredient Related disease Expression in TCGA** |
*the mean gene expression among soild cancers of TCGA | http://itcm.biotcm.net/detail/target.html?nId=11434 |
| Disease | English Name Definition External link |
http://itcm.biotcm.net/detail/disease.html?nId=6 |
the detailed page of ingredient is mainly composed of four parts: The part in the red box with "Profile--XX" is an brief introduction to the items.
The part in the orange box is a table of relationships (formula-herb+-ingredient-target-disease) about the items.
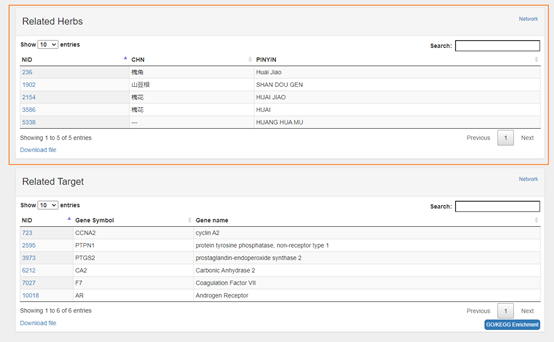
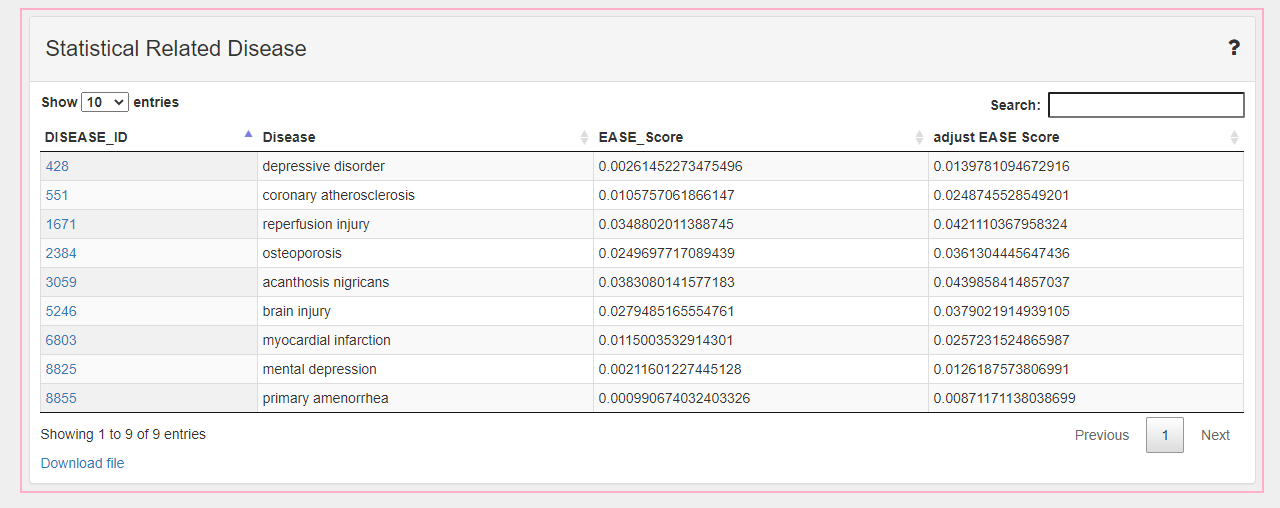
The part in the pink box with "Profile--XX" is a table of statistical relationship to the items.
notably, for a specific category, specific data will be displayed, for example, item in ingredient will display the properties and Potential Therapic Classification, or item in gene will display expression in TCGA.
In addition, on detailed page for each item, user can click the “network” button in the upper right corner of "related XXXX" part to a network diagram with item as the core.
View the detailed page for each item in SMEP module
Users can enter the details page from the 'SMEP' page by clicking the hyperlink of a certain item. the information cotained in item is summarized in the table below:
| source | Information contained | tips | Example link |
|---|---|---|---|
| Profile | Basic information DEGs visualization* Enrichment analysis** CMap Mapping GSEA*** GSVA*** Transcription Factor Analysis Tumor-related Pathway Analysis Immune-related Pathway Analysis |
*include Volcano plot and Heatmap **include KEGG/GO results in dotplots and tables ***signature curated form msigdb(C2/C3/C5/C7) |
http://itcm.biotcm.netdetail/profiledetail.html?nId=5 |
the detailed page of ingredient is mainly composed of two parts:
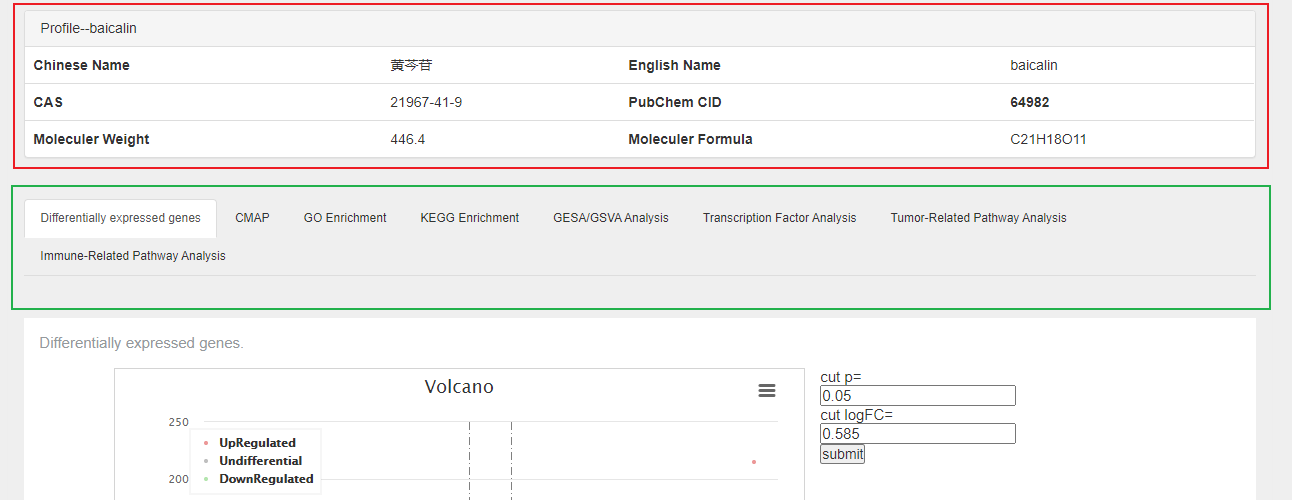
The part in the red box with "Profile--XX" is an brief introduction to the TCM ingredients, including name, cas, pubchem cid and so on.
The part in the green box with "Profile--XX" is collection of various analysis to the TCM ingredients, including differential expression analysis, functional enrichment analysis, connectivity map (CMap) analysis. and newest analysis, gene set enrichment analysis (GSEA), gene set variation analysis (GSVA), transcription factors enrichment analysis (TFEA).Users can click on the title and browse analysis results.
Explore the potential relationship between genesets and in TOOL module
Users can use algorithms deployed in the database by clicking the "TOOL" button on the navigation bar.
 '
'
There are three algorithms collected from advanced research and proved to be suitable for network pharmacology analysis and transcriptome analysis.
In this part, we briefly introduce how to use the algorithm, for more information about these algorithm, please read the Q&A part.
It may take a few minutes to get result, please be patient.
Download data
Information on CMTP in ITCM can be downloaded from the download page.
Information on SMEP in ITCM can be downloaded from the synapse (https://www.synapse.org/#!Synapse:syn25843090/files/).
Q&A
In ITCM, we use many new evaluation indexes and algorithms. We hope that the following questions can help users understand how it works and why we use it.
1. What is EASE Score used in Statistical Related Disease?
For Formula, Herb and Ingredient in CMTP MODULE, we use their related targets (Formula, Herb are generated by ingredients) and targets of diseases to predict statistical relationship by EASE Score.

In other TCM databases, the indirect relationship between ingredient and disease is evaluated by the Fisher’s exact test.
For example, for a specific disease term d and a specific ingredient i, the target union(n) regarding disease, formula, herb, ingredient are classified into four groups by the criteria: whether or not the targets are related to the ingredient, and whether or not the targets are related to diseases. Four numbers, a, b, c, and d are defined to represent the number of four category targets (see table below).
We then generated a 2×2 contingency table based on the four numbers and computed the P-value for the ingredient-disease pair i-d, P(i, d), using the following equation.
| e | in ingredient | not in ingredient | sum |
|---|---|---|---|
| in disease | a | b | a+b |
| not in disease | c | d | c+d |
| sum | a+c | b+d | n (=a+b+c+d) |
P(i,d)= ((a+b)!(c+d)!(a+c)!(b+d)!)/(a!b!c!d!n!)
However, we found that as long as there is an intersection between the ingredient and disease, significant differences will be obtained (P < 0.05)
Therefore, in order to make the statistical results more reliable, we decided to use EASE score as an index to evaluate the relationship between ingredients and diseases. the EASE Score is first introduced in DAVID[1] and is more conservative by subtracting one target from the both-in as seen below. If a = 1 or 0 (only one or none gene both in ingredient or disease), EASE Score is automatically set to 1.
| e | in ingredient | not in ingredient | sum |
|---|---|---|---|
| in disease | a-1 | b+1 | a+b |
| not in disease | c | d | c+d |
| sum | a+c-1 | b+d+1 | n (=a+b+c+d) |
EASE score for the ingredient-disease pair i-d, E(i,d) is computed as below:
E(i,d)= ((a+b)!(c+d)!(a+c-1)!(b+d+1)!)/((a-1)!(b+1)!c!d!n!)
Finlly, we collected relationships with E(i,d) <0.05 and adjust them by FDR-BH.
2. How do we get the properties of ingredients?
Tencent iDrug is adopted to get properties of ingredients.
Tencent iDrug (https://drug.ai.tencent.com/) is an AI-driven drug discovery platform that integrates molecular database, AI modelling and cloud computation, as well as workflow optimization, in one place.
more than 50 properties are generated by SMILES of ingredients and are classified into two groups: Basic properties and ADMET properties. Basic properties are related to the structure of ingredients, ADMET properties are predicted via multi-view graph neural networks developed in iDrug.
3. What is CMap mapping/CMap analysis?
The Connectivity Map[5], or CMap, is a powerful data resource that uses L1000 project transcriptional expression data to probe relationships between diseases, cell physiology, and therapeutics. it collected more than 30k profiles of small molecules treated with 9 cell lines. User can upload or enter their interested genesets into up-regulated genes and down-regulated genes box, and CMap will return a mapping result for reference perturbagen signatures (small molecules or mechanism of action) most similar (or dissimilar) in CMap touchstone dataset, in other words, CMap analysis is conducted to search drugs that may confer similar physiological effects. The results were quantified in normalized scores, from 100 to -100.
notably, there is a technical limit that number of genes in geneset is between 10-150. in our work, eligible TCM small molecule profile generate respective CMap mapping results and the clustering analysis is carried out to perform a wide pharmacological spectrum of TCM small molecules.
4. Which genesets/signatures are used in profile analysis?
Except GO enrichment analysis and CMap mapping, In our work, the reference gene sets to perform GSEA were curated from paper and Molecular Signatures Database (v7.5.1), and categorized as canonical pathway gene sets (KEGG, BIOCARTA, REACTOME, WikiPathways in subcollection C2), transcription factor target gene sets (GTRD in subcollection C3), MRNA target gene sets (miRDB in subcollection C3), hallmark gene sets (subcollection H), Metabolic genesets (paper). We performed GSEA in each gene sets respectively to get specific enrichment results and defined them as pathway analysis, miRNA analysis, transcription factor analysis, oncogenic pathway analysis and metabolic analysis.
5. What are the algorithms used in TOOL?
ITCM integrates a suite of existing and novel algorithms into an analysis environment. Here are the brief descriptions of algorithms/packages:
● SS Series (ss_xxxxx) are modified from R/Bioconductor packages for gene expression signature searching combined with functional enrichment analysis and visualization methods to facilitate the interpretation of the search results. in our work, the profile resources are high-throughput sequencing profiles of MCF-7 cells treated with 496 TCM small molecules, and the method CMap, lincs, fisher, corrlation, gCMap, ZhScore are tested and proven to be suitable to conduct transcriptional analysis on the above data.
6. Where can I find the gene expression data in SMEP?
The SMEP module collected 1488 high-throughput sequencing profiles of MCF-7 cells treated with 496 TCM small molecules. Each ingredient has 3 biological replicates. All the data are generated and curated by our team, and are available in the synapse (https://www.synapse.org/#!Synapse:syn25843090/files/).
Reference
[1] https://david.ncifcrf.gov/helps/functional_annotation.html#fisher
[2] Congreve M, Carr R, Murray C, Jhoti H. A 'rule of three' for fragment-based lead discovery?. Drug Discov Today. 2003;8(19):876-877.
[3] Chong, C. R., & Sullivan, D. J., Jr. New uses for old drugs. Nature, 2007;448(7154):645–646.
[4] Mao F, Ni W, Xu X, et al. Chemical Structure-Related Drug-Like Criteria of Global Approved Drugs. Molecules. 2016;21(1):75.
[5] Subramanian A, et al. A Next Generation Connectivity Map: L1000 Platform And The First 1,000,000 Profiles. Cell. 2017/12/1. 171(6):1437–1452.
[6] Valdeolivas A, Tichit L, Navarro C, et al. Random walk with restart on multiplex and heterogeneous biological networks. Bioinformatics. 2019;35(3):497-505.
[7] Peng Y, Yuan M, Xin J, Liu X, Wang J. Screening novel drug candidates for Alzheimer's disease by an integrated network and transcriptome analysis. Bioinformatics. 2020;36(17):4626-4632.